An Open Science Approach to Machine Learning in Biomedical Research
Batool Almarzouq- @batool664
A little bit about me!
- A computational biologist affiliated with the University of Liverpool.
- Founder of RLadies Chapter in Saudi Arabia (Dammam).
- A curator in the R Weekly team.
- Member of MiR accessibility committee.
- Member in the turing way community.
- Working on establishing an Open Science community in Saudi Arabia.
Acknowledgment
- Anelda Van der
- Malvika Sharan, Kirstie Whitaker and Martina G. Vilas
- The Turing Way Community
- Alison Presmanes Hill (slides)
Why do we use ML in Biomedical Research?
Image Credit: ABC Science
Image Credit: ABC Science
DNA
- DNA sequence alignment
- DNA sequence classification
- DNA sequence clustering
- DNA pattern mining
Algorithms includes fuzzy sets, neural networks, genetic algorithms.
Image Credit: ABC Science
DNA
- DNA sequence alignment
- DNA sequence classification
- DNA sequence clustering
- DNA pattern mining
Algorithms includes fuzzy sets, neural networks, genetic algorithms.
RNA
- Mainly RNA-sequencing (RNA-seq)
- Differentially expressed genes (DEGs)
- Alternative splicing
- Small RNA expression
Algorithms include Logistic Regression, Random Forest, LMT, Random Subspace.
Solving the sequence is not enough!
We need to know the structure and function of the protein!
Image Credit: doi:10.1021/cr400525m
Image Credit: doi:10.1021/cr400525m
How can we predict function from structure?
To predict the function from the structure, scientists use different approaches including machine learning (ML) and deep learning algorithms .
Credit: Supriyo Bhattacharya/Beckman Research Institute at City of Hope
Credit: Supriyo Bhattacharya/Beckman Research Institute at City of Hope
Prediction of protein structure is important to develop small molecules and targeted therapy for diseases.
Why not only rely on Experemtal Methods?
Credit: Data for UniProtKB obtained form Claire O'Donovan via EBI database support
Credit: Data for UniProtKB obtained form Claire O'Donovan via EBI database support
Because of the growing gap between the newly-sequenced and characterized sequences in the genome databases, computational methods in gene functional annotation are indispensable. Moreover, given the drop in the genome sequencing techniques' cost, this gap is only destined to grow.
Biology has become a highly data-intensive science, dependent on complex, computational, and statistical methods!
So, how can we make these methods available and accessible for researchers, while ensuring that scientific results remain reproducible?
What is the percentage of reproducible research?
Credit: Key results of the survey on reproducibility conducted by Nature in 2016
How can we overcome the reproducibility crisis?
How can you improve the reproducibility of your data science project?
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
Share Computational ENVIRONMENT
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
Share Computational ENVIRONMENT
VERSION CONTROL
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
Share Computational ENVIRONMENT
VERSION CONTROL
TESTING
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
Share Computational ENVIRONMENT
VERSION CONTROL
TESTING
DOCUMENTATION
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
Share Computational ENVIRONMENT
VERSION CONTROL
TESTING
DOCUMENTATION
OPEN DATA/FAIR DATA
How can you improve the reproducibility of your data science project?
OPEN SOURCE SOFTWARE
SHARE CODE/ANALYSIS
Share Computational ENVIRONMENT
VERSION CONTROL
TESTING
DOCUMENTATION
OPEN DATA/FAIR DATA
OPEN ACCESS
This is called Open Science.
Open Science is about extending the principles of openness to the whole research cycle, fostering sharing and collaboration as early as possible thus entailing a systemic change to the way science and research is done
-- FOSTER Plus
What are the FAIR principles?
* The Turing Way project illustration by Scriberia. Zenodo. http://doi.org/10.5281/zenodo.3332807
Why do we use version control (git)?
Version Control in the Old Days ..


Real Version Control (including backup)
In the pandemic, some publishers have “opened” their journals to make certain articles freely available.
In the pandemic, some publishers have “opened” their journals to make certain articles freely available.
Databases have been created that are completely open access, such as the Open COVID Pledge.
UNESCO is launching international consultations aimed at developing a Recommendation on Open Science for adoption by member states in 2021
UNESCO is launching international consultations aimed at developing a Recommendation on Open Science for adoption by member states in 2021
There is a network of Open Science Communities in Netherlands, Sweden, Germany, UK and others
In line with vision 2030, we are starting an Open Science Community in Saudi Arabia.
It's created and developed with the help of the "Open Life Sciences"

Open Life Sciences (OLS3) program helps individuals and stakeholders in research to become Open Science ambassadors.
We want to provide a place where newcomers and experienced peers interact, inspire each other to embed open science (research) practices and values in their workflows and provide feedback on policies, infrastructures and support services. Together working to make Open Science the norm. So we are calling out to researchers and colleagues in Saudi Arabia.
 Batool Almarzouq
The University of Liverpool
Batool Almarzouq
The University of Liverpool
 Founder and director of Talarify, Mentor OLS3
Founder and director of Talarify, Mentor OLS3
 Paula Moraga, Assistant Professor in Statistics for Public Health
(KAUST)
Paula Moraga, Assistant Professor in Statistics for Public Health
(KAUST)
Join me on the 24th of Feb for a workshop titled "Collaborating on Open Data Science Projects" as part of the Datathon for WiDS2021.
How can you start learning about Open Science?
How can you start learning about Open Science?
* The Turing Way project illustration by Scriberia. Zenodo. http://doi.org/10.5281/zenodo.3332807
Kirstie Whitaker, Project Lead

Malvika Sharan, Community Manager

So, What is the turing way?
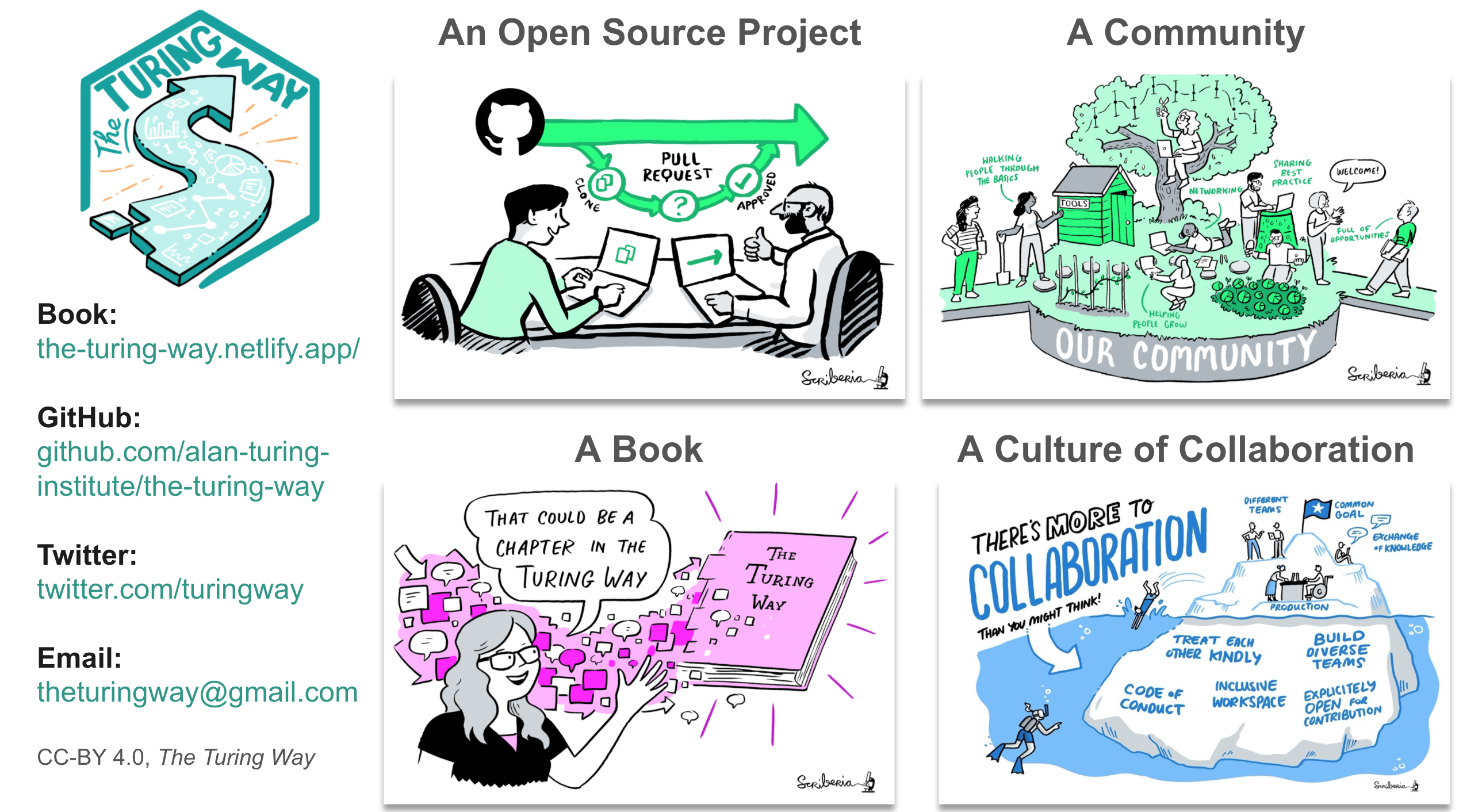

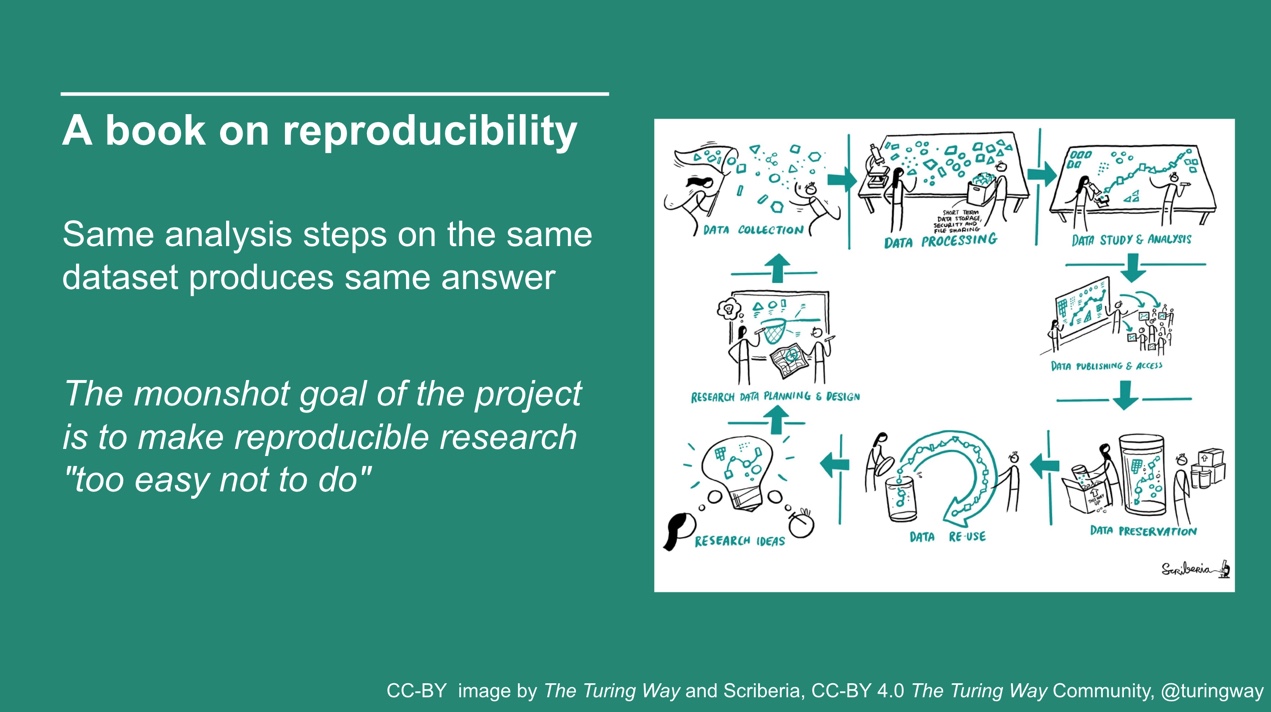
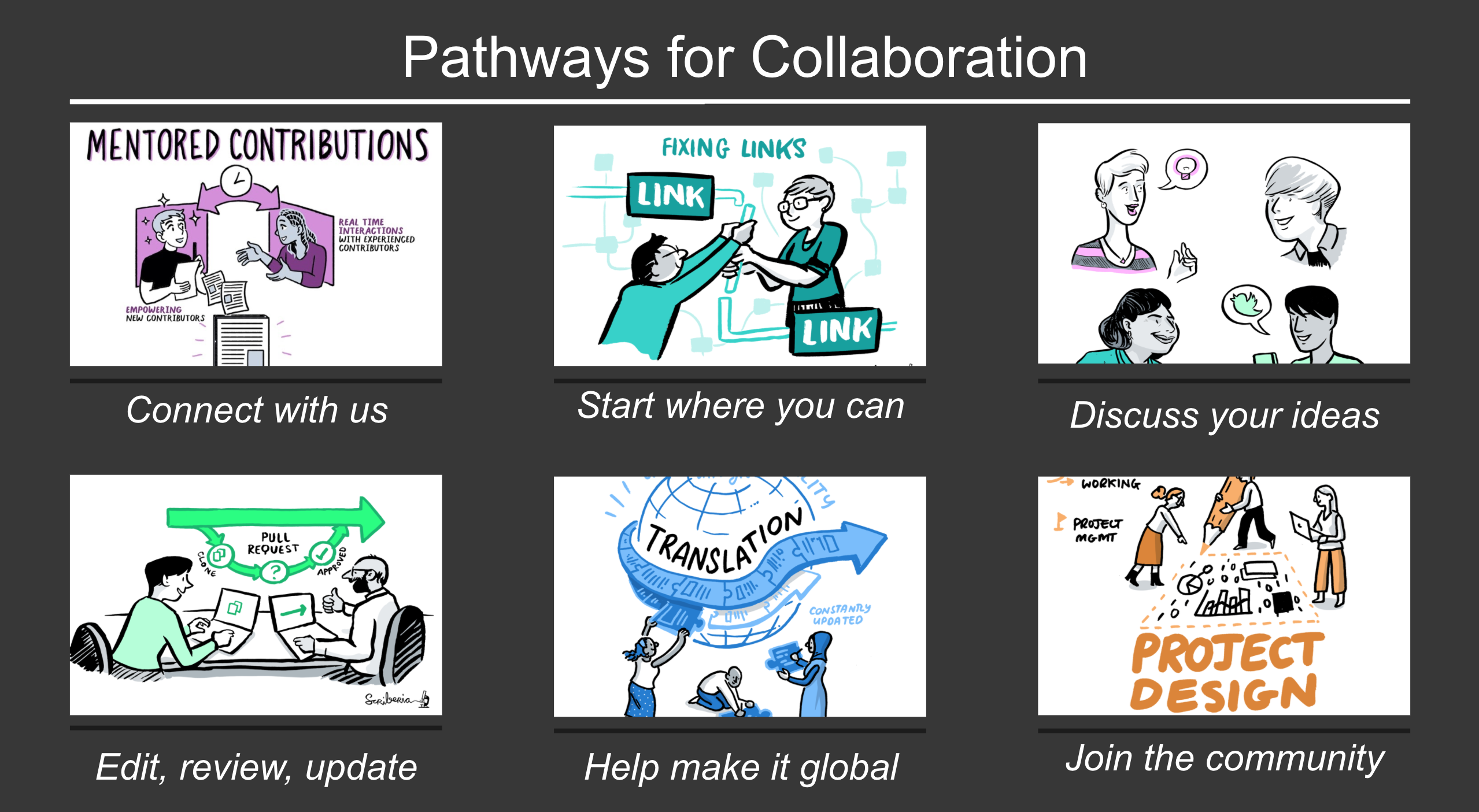
Join the next book dash event!
Book Dash November 2020
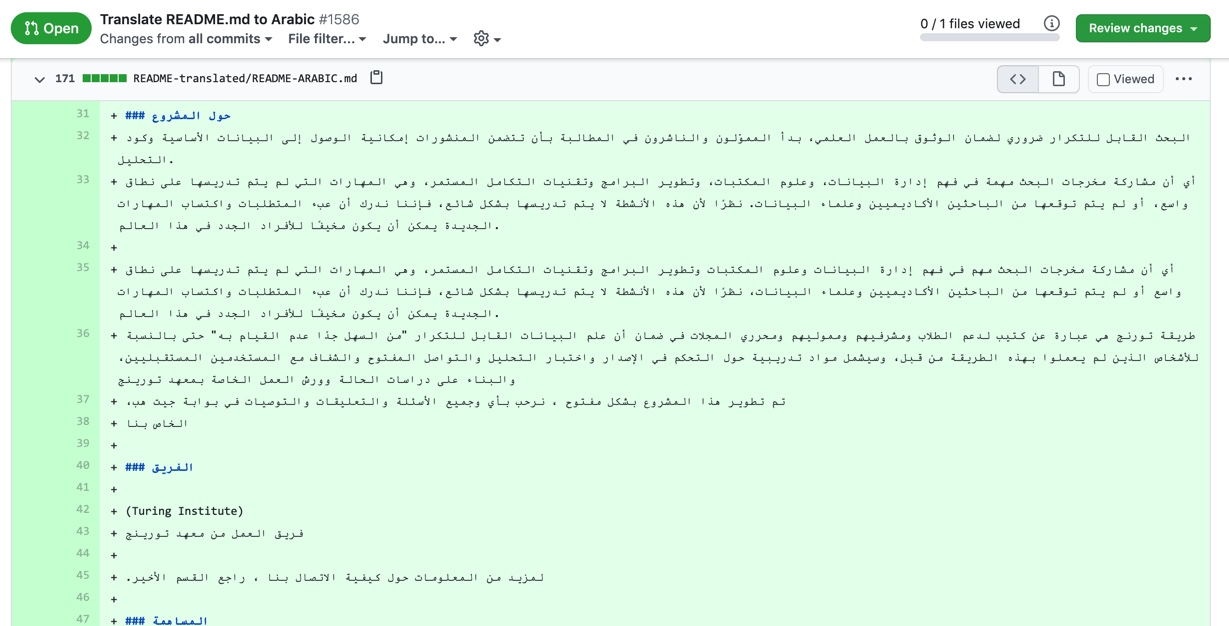
Review README.md Arabic translation
Upcoming Workshop by the turing way
Register for the free workshop 'Boost your research reproducibility with Binder' run by Sarah Gibson from the Turing Way as part of our Research Software Camp on research accessibility.
